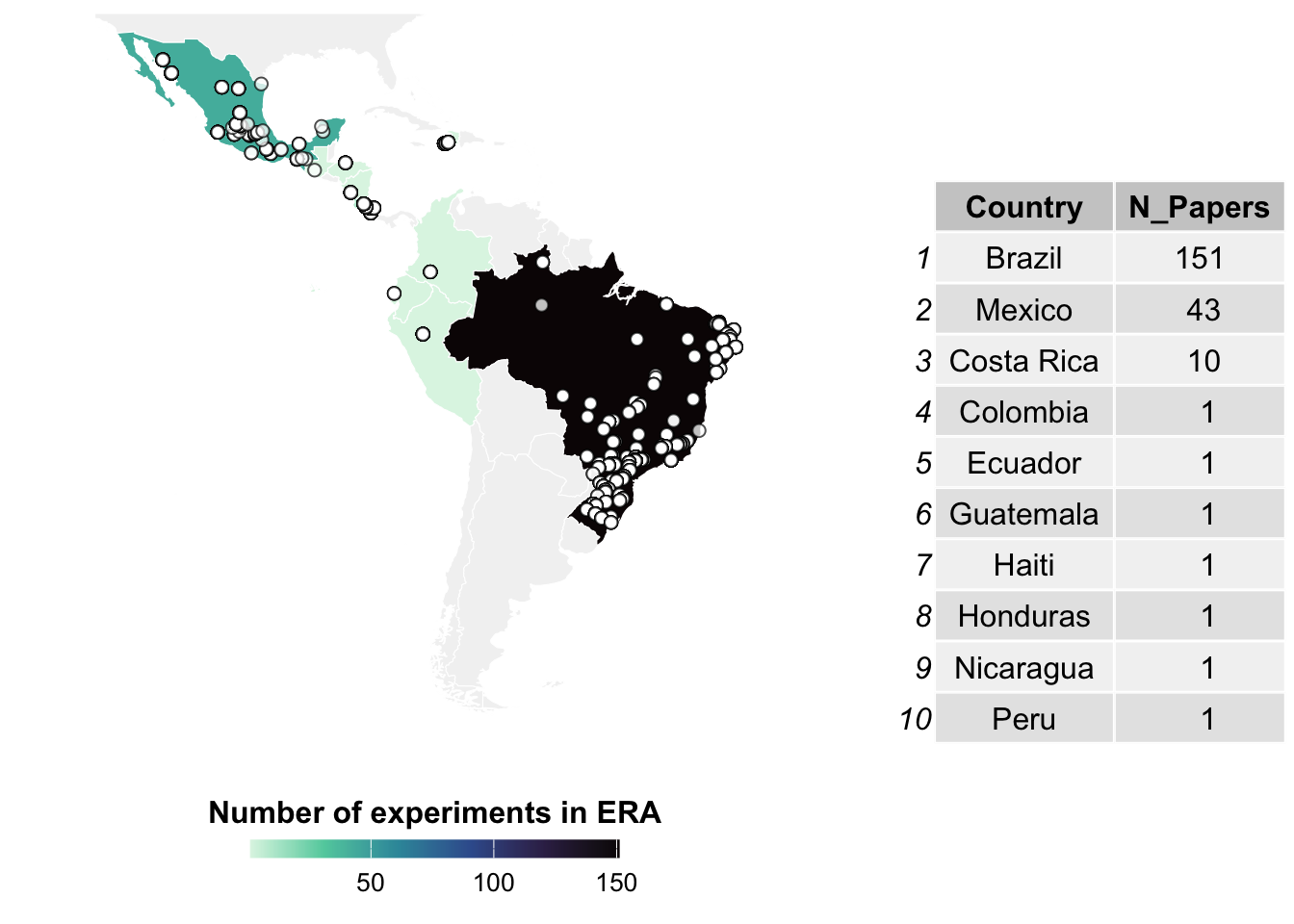
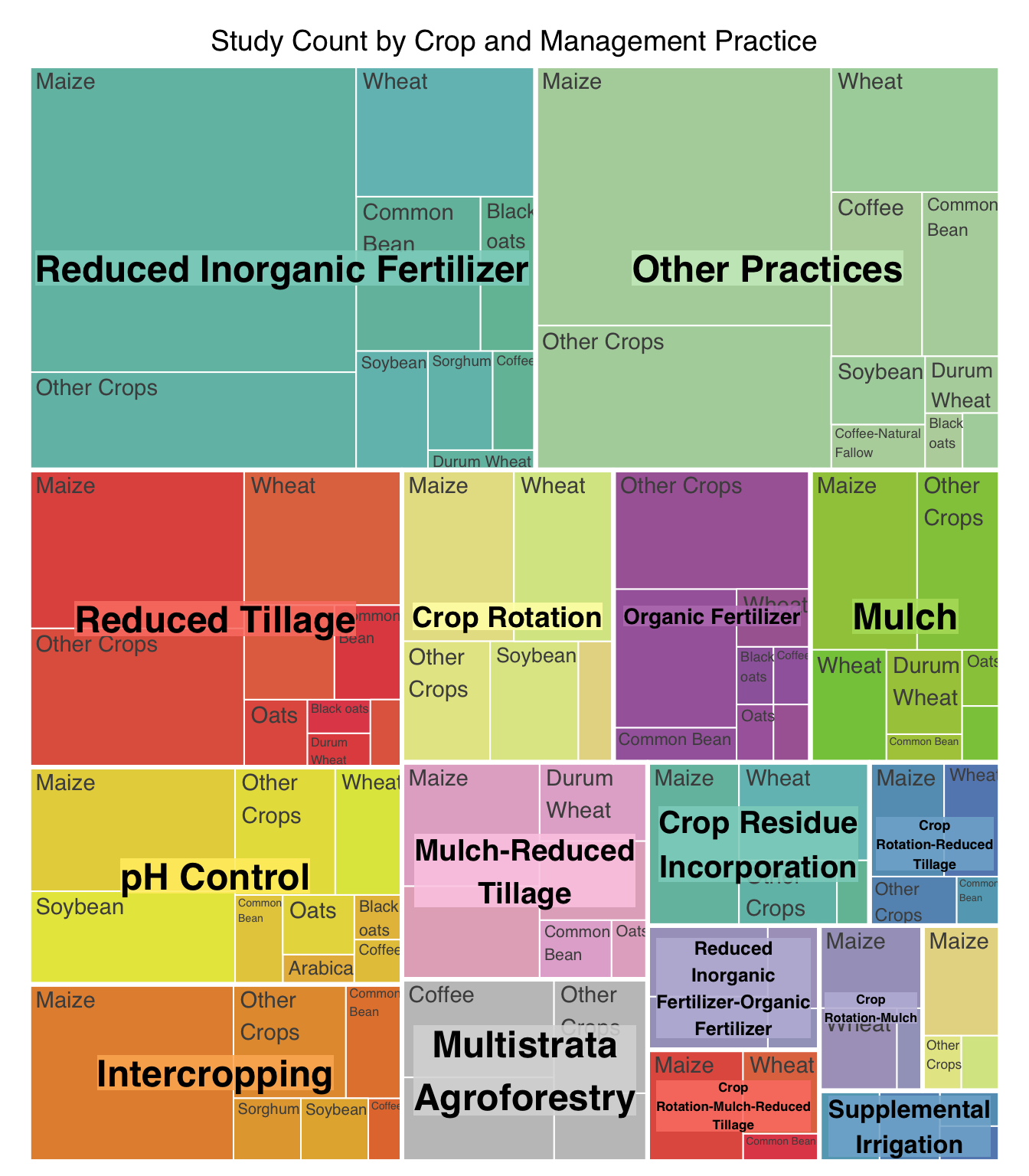
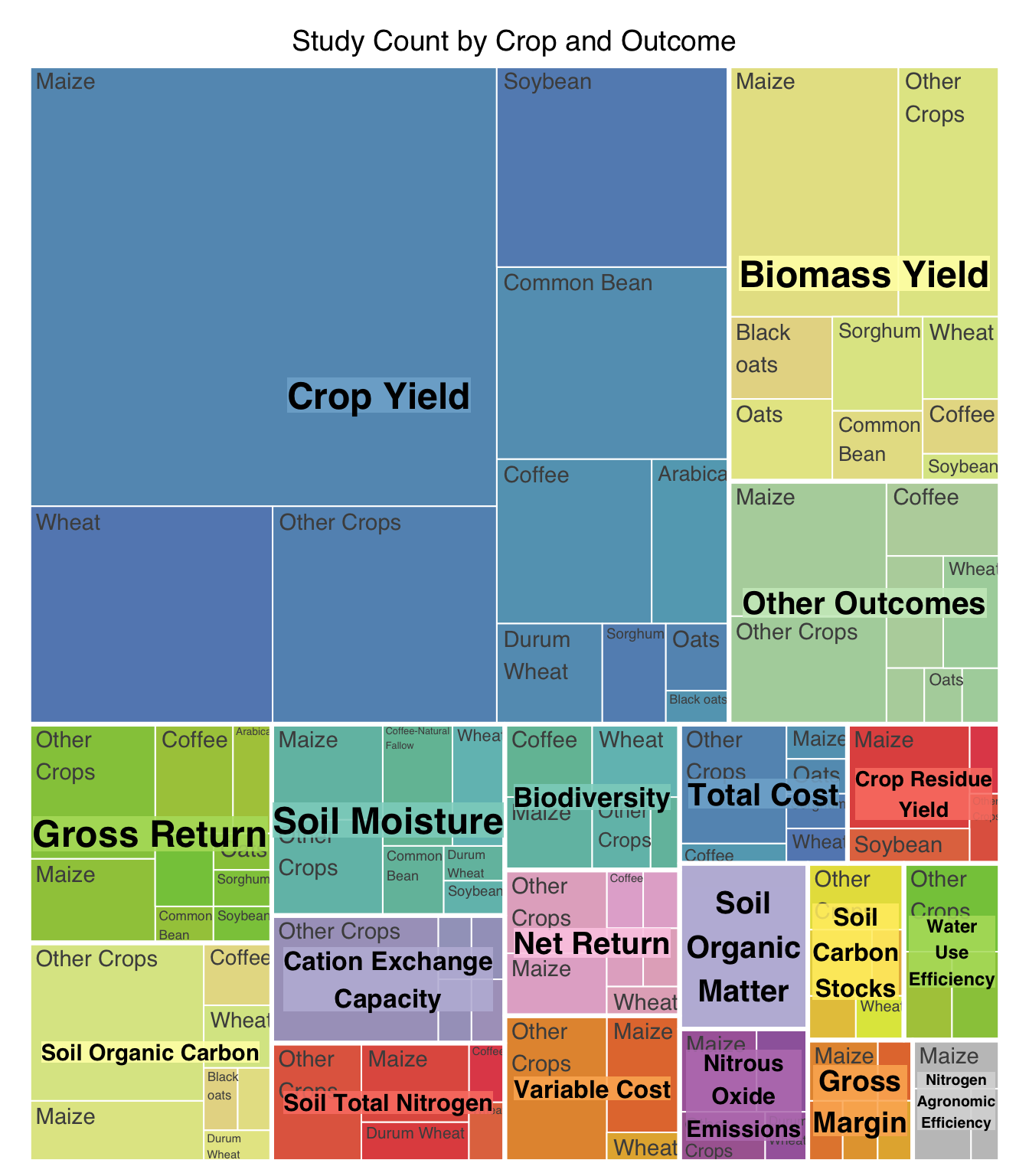
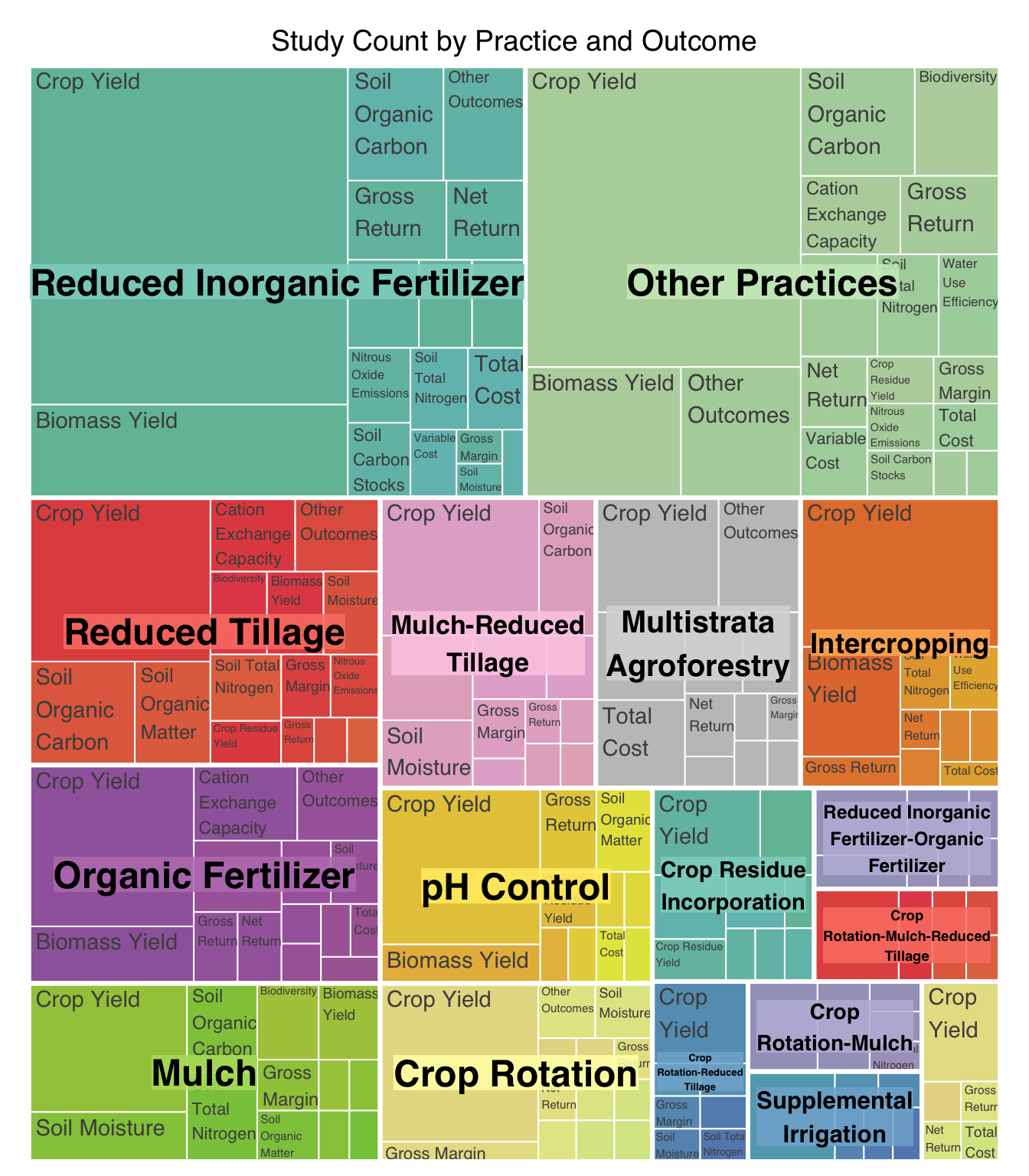
| Reduced Inorganic Fertilizer |
82 |
2531 |
| Reduced Tillage |
32 |
5065 |
| Intercropping |
27 |
302 |
| Organic Fertilizer |
22 |
389 |
| Crop Rotation |
20 |
982 |
| Mulch-Reduced Tillage |
18 |
584 |
| Mulch |
18 |
968 |
| pH Control |
17 |
874 |
| Multistrata Agroforestry |
15 |
904 |
| Crop Residue Incorporation |
10 |
421 |
| Reduced Inorganic Fertilizer-Organic Fertilizer |
8 |
76 |
| Crop Rotation-Reduced Tillage |
7 |
51 |
| Crop Rotation-Mulch-Reduced Tillage |
6 |
74 |
| Supplemental Irrigation |
5 |
231 |
| Reduced Inorganic Fertilizer-Intercropping |
5 |
64 |
| Crop Rotation-Mulch |
5 |
54 |
| Crop Rotation-Reduced Inorganic Fertilizer |
4 |
16 |
| Crop Residue Incorporation-Crop Rotation |
4 |
38 |
| Improved Fallow |
3 |
182 |
| Reduced Inorganic Fertilizer-pH Control |
3 |
154 |
| Other Agroforestry |
3 |
64 |
| Crop Residue Incorporation-Reduced Tillage |
3 |
15 |
| Improved Varieties |
3 |
279 |
| Reduced Inorganic Fertilizer-Mulch-Reduced Tillage |
3 |
78 |
| Reduced Inorganic Fertilizer-Mulch |
3 |
111 |
| Crop Residue |
3 |
242 |
| Crop Residue Incorporation-Mulch-Reduced Tillage |
2 |
28 |
| Improved Fallow-Intercropping |
2 |
240 |
| Double Cropping |
2 |
17 |
| Improved Varieties-Intercropping |
2 |
10 |
| Deficit Irrigation |
2 |
178 |
| Biochar |
2 |
76 |
| Alleycropping |
2 |
396 |
| Agroforestry Pruning |
2 |
32 |
| Biochar-Organic Fertilizer |
2 |
31 |
| Crop Residue-Reduced Tillage |
2 |
42 |
| Crop Residue-Intercropping-Reduced Tillage |
2 |
8 |
| Agroforestry Pruning-Multistrata Agroforestry |
2 |
19 |
| Green Manure |
2 |
27 |
| Harvest Method-Intercropping-Mulch |
1 |
21 |
| Crop Rotation-Intercropping-Reduced Tillage |
1 |
1 |
| Intercropping-Supplemental Irrigation |
1 |
1 |
| Improved Fallow-Reduced Inorganic Fertilizer |
1 |
2 |
| Improved Fallow-Reduced Inorganic Fertilizer-Intercropping |
1 |
2 |
| Agroforestry Pruning-Mulch |
1 |
8 |
| Agroforestry Pruning-Mulch-pH Control |
1 |
4 |
| Improved Varieties-pH Control |
1 |
30 |
| Improved Varieties-Reduced Inorganic Fertilizer-pH Control |
1 |
120 |
| Intercropping-Mulch |
1 |
14 |
| Multistrata Agroforestry-Supplemental Irrigation |
1 |
26 |
| Reduced Inorganic Fertilizer-Multistrata Agroforestry-pH Control |
1 |
142 |
| Reduced Inorganic Fertilizer-Multistrata Agroforestry |
1 |
288 |
| Biochar-Reduced Inorganic Fertilizer-Organic Fertilizer |
1 |
10 |
| Crop Rotation-Reduced Inorganic Fertilizer-Mulch |
1 |
3 |
| Crop Residue Incorporation-Crop Rotation-Mulch-Reduced Tillage |
1 |
12 |
| Organic Fertilizer-pH Control |
1 |
16 |
| Improved Varieties-Multistrata Agroforestry |
1 |
8 |
| Mulch-pH Control |
1 |
3 |
| Crop Rotation-pH Control |
1 |
4 |
| pH Control-Reduced Tillage |
1 |
4 |
| Alleycropping-Crop Rotation |
1 |
4 |
| Crop Rotation-Intercropping |
1 |
8 |
| Crop Rotation-Reduced Inorganic Fertilizer-Intercropping |
1 |
4 |
| Parklands |
1 |
4 |
| Mulch-Organic Fertilizer |
1 |
2 |
| Agroforestry Pruning-Alleycropping |
1 |
48 |
| Agroforestry Pruning-Alleycropping-Reduced Inorganic Fertilizer |
1 |
24 |
| Agroforestry Pruning-Organic Fertilizer |
1 |
30 |
| Crop Residue Incorporation-Mulch |
1 |
26 |
| Crop Residue-Intercropping |
1 |
2 |
| Other Agroforestry-Parklands |
1 |
95 |
| Intercropping-Reduced Tillage |
1 |
4 |
| Improved Varieties-Reduced Inorganic Fertilizer-Intercropping |
1 |
6 |
| Agroforestry Pruning-Reduced Inorganic Fertilizer |
1 |
18 |
| Crop Residue-Crop Rotation-Reduced Tillage |
1 |
10 |
| Biochar-Mulch-Organic Fertilizer |
1 |
3 |
| Biochar-Reduced Inorganic Fertilizer-Mulch-Organic Fertilizer |
1 |
1 |
| Crop Residue-Mulch |
1 |
76 |
| Crop Residue Incorporation-Reduced Inorganic Fertilizer |
1 |
6 |
| Reduced Inorganic Fertilizer-Reduced Tillage |
1 |
4 |
| Crop Residue-Mulch-Reduced Tillage |
1 |
36 |
| Agroforestry Pruning-Reduced Tillage |
1 |
2 |
| Green Manure-Reduced Inorganic Fertilizer |
1 |
3 |
| Green Manure-Organic Fertilizer |
1 |
3 |
| Green Manure-Reduced Inorganic Fertilizer-Organic Fertilizer |
1 |
2 |
| Crop Rotation-Organic Fertilizer |
1 |
3 |
| Crop Rotation-Reduced Inorganic Fertilizer-Organic Fertilizer |
1 |
2 |